Introduction
Rare diseases affect over 350 million people worldwide, 50% of which are children [1, 2]. Because each disease is rare, a general physician may have never seen a patient with a particular disease in their entire career [3]. Symptoms can also vary considerably between individual cases, making correct diagnosis difficult [4]. Misdiagnosis and incorrect treatment are extremely costly and potentially dangerous, so it is important to quickly refer patients to specialists that are experts in their condition. To facilitate this referral process, we trained machine learning models to predict the expertise of each researcher in each rare disease, based on their publication record. We compared the performance of three methods on a dataset of 209,110 disease-author associations, and were able to classify rare disease experts from GeneReviews with 76% accuracy and predict 21,224 new disease-expert associations.
Approach
We hypothesized that it is possible to distinguish experts from non-experts in a particular disease based on their publication record for that disease. The more frequently a researcher publishes on a disease, the more likely they are to be an expert.
To test this hypothesis, we compiled a data set of 2,160 known disease-expert associations and 206,950 unknown disease-author associations, based on the authorship of publications associated with rare diseases (see Figure 1). Rather than attempt to identify whether or not a disease is the topic of an article based on its abstract, we obtained a curated list of associated publications for each rare disease from OMIM, an online catalog of rare inherited diseases [5]. Known disease-expert associations were obtained from GeneReviews, a resource with high-quality peer-reviewed summaries of a variety of inherited conditions [6]. GeneReviews chapters are written by one or more experts and focus on a specific condition or disease. The positive disease-expert associations from GeneReviews publications were combined with the unlabeled disease-author associations from OMIM to form the complete data set.
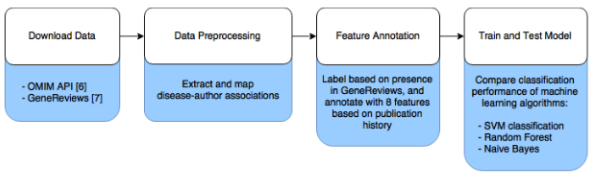
Figure 1: Data analysis workflow.
To combine these data sets, author names needed to be extracted from both OMIM references and GeneReviews articles and then merged, and GeneReviews articles needed to be linked to diseases in OMIM. The formatting of author names differed between OMIM and GeneReviews, so we used the combination of the full last name, first initial, and middle initial (if present) to uniquely identify an author. We identified 98,705 unique authors, with 1,724 having published in both GeneReviews and articles included in OMIM. GeneReviews chapters frequently include a table of OMIM identifiers, with 95% of the 664 chapters we analyzed linking to one or more OMIM records. The GeneReview authors were considered to be experts in each linked OMIM record if they had at least one other publication associated with that OMIM record. Unlabeled disease-author associations were taken from the 1,292 OMIM records with at least one associated GeneReview article. OMIM records can correspond to either diseases or disease-associated genes, but we did not distinguish these cases.
| Model | Precision | Recall | Accuracy | F1 |
|---|---|---|---|---|
| SVM | 0.76 | 0.76 | 0.76 | 0.76 |
| Random Forest | 0.72 | 0.78 | 0.74 | 0.75 |
| Naive Bayes | 0.75 | 0.65 | 0.72 | 0.70 |
Table 1: The classification performance of different learning algorithms
We designed 8 features to discriminate the disease-author associations of experts from non-experts: 1) number of publications by the author on the disease, 2) standardized number of publications by the author on the disease (zero mean and unit variance across authors per disease), 3) number of diseases that the author has published on, 4) number of first-author publications by the author on the disease, 5) number of last-author publications by the author on the disease, and the number of publications by the author on the disease in the last 6) 3 years, 7) 5 years, and 8) 10 years.
We trained three learning algorithms, an SVM classifier [7], Random Forest [8] and naive Bayes [9], on the 2,160 positive disease-expert associations and 2,160 randomly-selected unlabeled disease-author associations which served as negative examples. We averaged the performance across ten separate iterations of 10-fold cross-validation, each with a different randomly selected set of negative examples.
Analysis
The SVM classifier outperformed the other methods in accuracy, precision, and F1 measure, with both precision and recall of 76% (see Table 1). The Random Forest and naive Bayes models performed slightly worse, though the naive Bayes model achieved higher recall than the SVM (78%).
Most disease-author associations were confidently classified by the SVM, and misclassified cases were often confidently misclassified (see Figure 2). While the SVM classifier scored most negative disease-author associations below 0.2, some associations were scored very highly. We used unlabeled associations as negative examples under the assumption that most authors that have published on a disease are not experts in that disease. However, a substantial fraction of these unlabeled associations will represent real expertise where the author has simply not published in GeneReviews. The classifier predicted 21,224 disease-expert associations out of 206,950 unlabeled associations (10%).
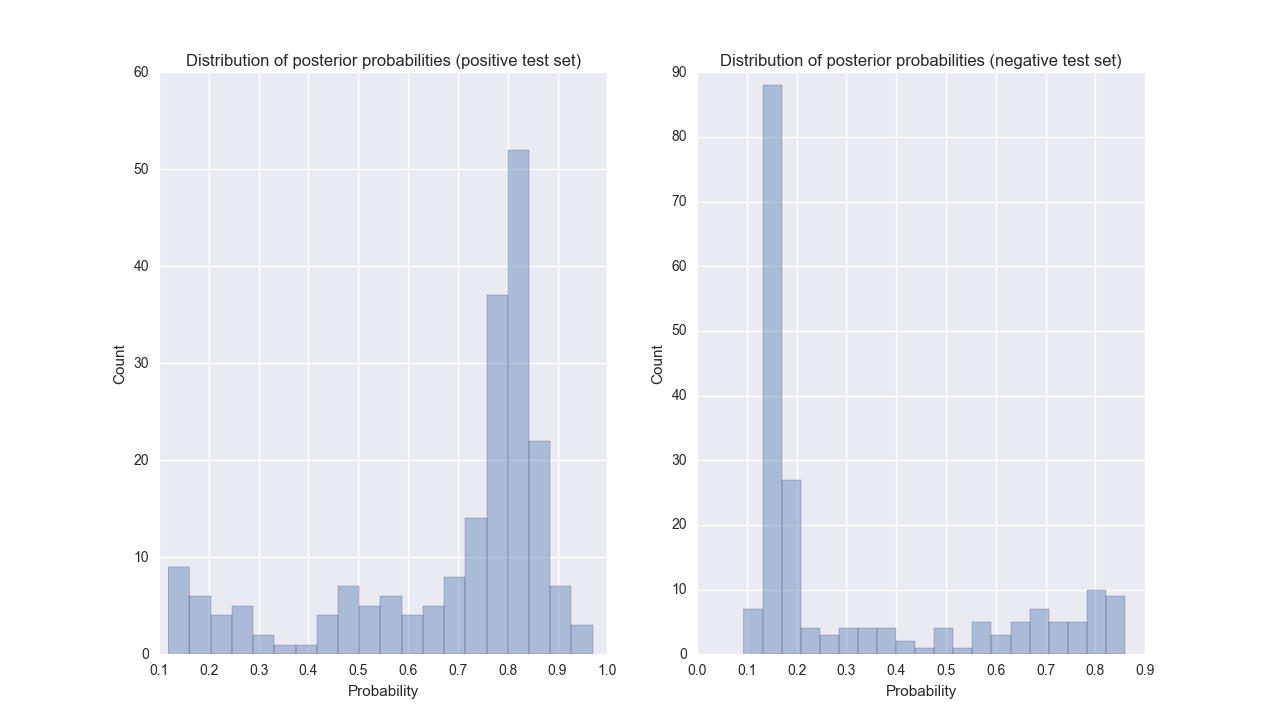
Figure 2: Distributions of the posterior probability from the SVM classifier for positive (left) and negative (right) test data during one iteration of cross-validation.
Conclusion
Using an SVM model, we were able to accurately predict the disease expertise of researchers based on features extracted from their publication history. This method can improve the referral of patients to rare disease specialists, thereby reducing the time to eventual diagnosis. Additionally, this method can be used to suggest journal reviewers or conference invitees where expertise in a specific disease is desired. In future work, we plan to incorporate author disambiguation to improve the accuracy of the author publication history. This work also depends on the accuracy and completeness of OMIM’s curated references, which could be augmented by using text mining to automatically identify publications associated with rare diseases.
References
-
Rare Disease Day 2016 - 29 Feb - Article.http://www.rarediseaseday.org/article/what-is-a-rare-disease. Accessed: 2016-08-22.
-
J. Mendlovic, H. Barash, H. Yardeni, Y. Banet-Levi, H. Yonath, and A. Raas-Rothschild. Rare diseases DTC: Diagnosis, treatment and care. Harefuah,155(4):241–253, 2016.
-
Rare diseases strategy - Publications - GOV.UK.https://www.gov.uk/government/publications/rare-diseases-strategy. Accessed: 2016-08-22.
-
H. Singh, T. Giardina, A. Meyer, S. Forjuoh, M. Reis, and E. Thomas. Types and origins of diagnostic errors in primary care settings. JAMA Internal Medicine, 173(6):418, 2013.
-
Joanna S Amberger, Carol A Bocchini, Franc ̧ois Schiettecatte, Alan F Scott, andAda Hamosh. OMIM.org: Online Mendelian Inheritance in Man (OMIMR©), anonline catalog of human genes and genetic disorders. Nucleic acids research, 43(D1):D789–D798, 2015
-
Roberta A Pagon. GeneReviews. University of Washington, 1993.
-
Ting-Fan Wu, Chih-Jen Lin, and Ruby C Weng. Probability estimates formulti-class classification by pairwise coupling. Journal of Machine Learning Research, 5(Aug):975–1005, 2004.
-
Leo Breiman. Random forests. Machine learning, 45(1):5–32, 2001.
-
Harry Zhang. Exploring conditions for the optimality of naive Bayes. International Journal of Pattern Recognition and Artificial Intelligence, 19(02):183–198, 2005.